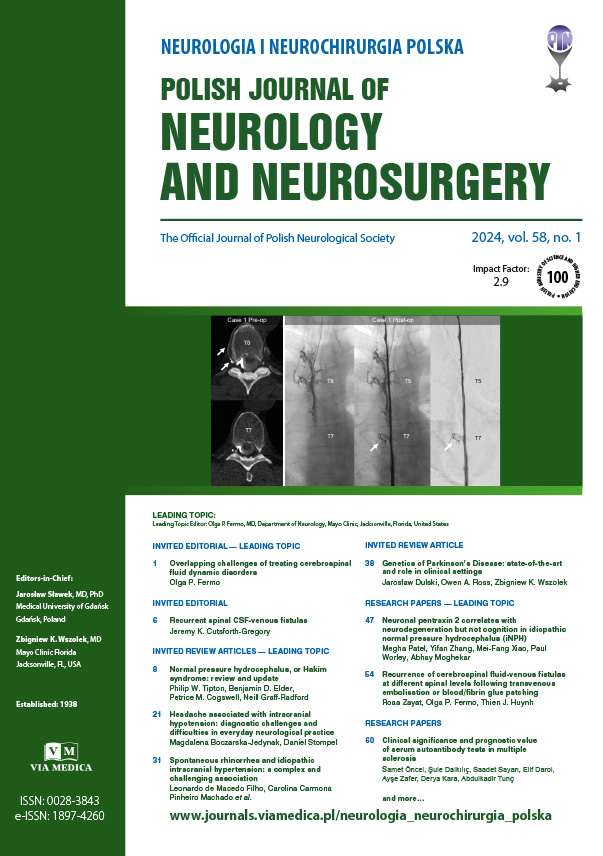
Welcome to Via Medica Journals’ Service!
Since 1993 Via Medica has been publishing journals in various fields of medicine, both clinical and experimental. Nowadays it is more than 40 titles, which include scientific journals published in English and addressed to professionals worldwide. Our portfolio also contains educational journals for readers in Poland, especially for young physicians during the residency. Many of these titles are published in close cooperation with various Polish medical societies, what appropriate level of expertise and facilitates further development. Most of our journals provide free access to all archival resources. Several of them are fully available in open access, which is part of the idea of free access to knowledge and the dissemination of science. Our experts contribute to constant, both operational and conceptual development of journals published by Via Medica — always in direct cooperation with Editorial Offices. We support processes of improving quality of editorial tools, indexing journals in the Polish and worldwide scientific databases as well as provide journals’ advanced scientometric analyses based upon international guidelines and application of well-established bibliometric indicators. Moreover, we coordinate the optimization of our manuscript system, are involved in the promotion of Via Medica periodicals and also in implementation of new medical journals.
View